Automated image classification is a promising branch of machine learning (ML) useful for skin cancer diagnosis, but little has been determined about its limitations for general usability in current clinical practice.
ObjectiveTo determine limitations in the selection of skin cancer images for ML analysis, particularly in melanoma.
MethodsRetrospective cohort study design, including 2,849 consecutive high-quality dermoscopy images of skin tumors from 2010 to 2014, for evaluation by a ML system. Each dermoscopy image was assorted according to its eligibility for ML analysis.
ResultsOf the 2,849 images chosen from our database, 968 (34%) met the inclusion criteria for analysis by the ML system. Only 64.7% of nevi and 36.6% of melanoma met the inclusion criteria. Of the 528 melanomas, 335 (63.4%) were excluded. An absence of normal surrounding skin (40.5% of all melanomas from our database) and absence of pigmentation (14.2%) were the most common reasons for exclusion from ML analysis.
DiscussionOnly 36.6% of our melanomas were admissible for analysis by state-of-the-art ML systems. We conclude that future ML systems should be trained on larger datasets which include relevant non-ideal images from lesions evaluated in real clinical practice. Fortunately, many of these limitations are being overcome by the scientific community as recent works show.
La clasificación automática de imágenes es una rama prometedora del aprendizaje automático (de sus siglas en inglés Machine Learning [ML]), y es una herramienta útil en el diagnóstico de cáncer de piel. Sin embargo, poco se ha estudiado acerca de las limitaciones de su uso en la práctica clínica diaria.
ObjetivoDeterminar las limitaciones que existen en cuanto a la selección de imágenes usadas para el análisis por ML de las neoplasias cutáneas, en particular del melanoma.
MétodosSe diseñó un estudio de cohorte retrospectivo, donde se incluyeron de forma consecutiva 2.849 imágenes dermatoscópicas de alta calidad de tumores cutáneos para su valoración por un sistema de ML, recogidas entre los años 2010 y 2014. Cada imagen dermatoscópica fue clasificada según las características de elegibilidad para el análisis por ML.
ResultadosDe las 2.849 imágenes elegidas a partir de nuestra base de datos, 968 (34%) cumplieron los criterios de inclusión. De los 528 melanomas, 335 (63,4%) fueron excluidos. La ausencia de piel normal circundante (40,5% de todos los melanomas de nuestra base de datos) y la ausencia de pigmentación (14,2%) fueron las causas más frecuentes de exclusión para el análisis por ML.
DiscusiónSolo el 36,6% de nuestros melanomas se consideraron aceptables para el análisis por sistemas de ML de última generación. Concluimos que los futuros sistemas de ML deberán ser entrenados a partir de bases de datos más grandes que incluyan imágenes representativas de la práctica clínica habitual. Afortunadamente, muchas de estas limitaciones están siendo superadas gracias a los avances realizados recientemente por la comunidad científica, como se ha demostrado en trabajos recientes.
Automated image classification by pattern recognition is a branch of machine learning (ML) which offers the dermatologist a useful tool for assessment in the diagnosis of skin cancer.1 Deep convolutional neural networks (DCNN) have dramatically improved accuracy in feature learning and object classification2 and have been successfully used in the classification of dermoscopic images of skin lesions.3 However, the selection of images may include certain special features which prevent its universal use at the present time. In this study we assessed some exclusion criteria in the selection of skin cancer images (with an emphasis on melanoma) for ML analysis, according to recent works in this field.1,4,5
Materials and MethodsThis study was conducted in a tertiary academic skin cancer center in Barcelona, Spain. A retrospective cohort study was designed including 2,849 consecutive high-quality dermoscopy images of skin tumors from the Melanoma Unit database from 2010 to 2014. The DermLite® photo digital epiluminescence microscopy system 3Gen with 37mm thread size and a Canon camera, model G16 were used. Pathological diagnosis was available for 2,429 images. Finally, the images were assorted according to their theoretical eligibility for ML analysis, pursuant to some potential exclusion criteria1,4,5: difficulty in lesion border detection (absence of pigmentation, absence of normal surrounding skin, presence of hair, location on volar skin), metastasis or an ulcerated lesion.
This study has been approved by the institutional review board. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
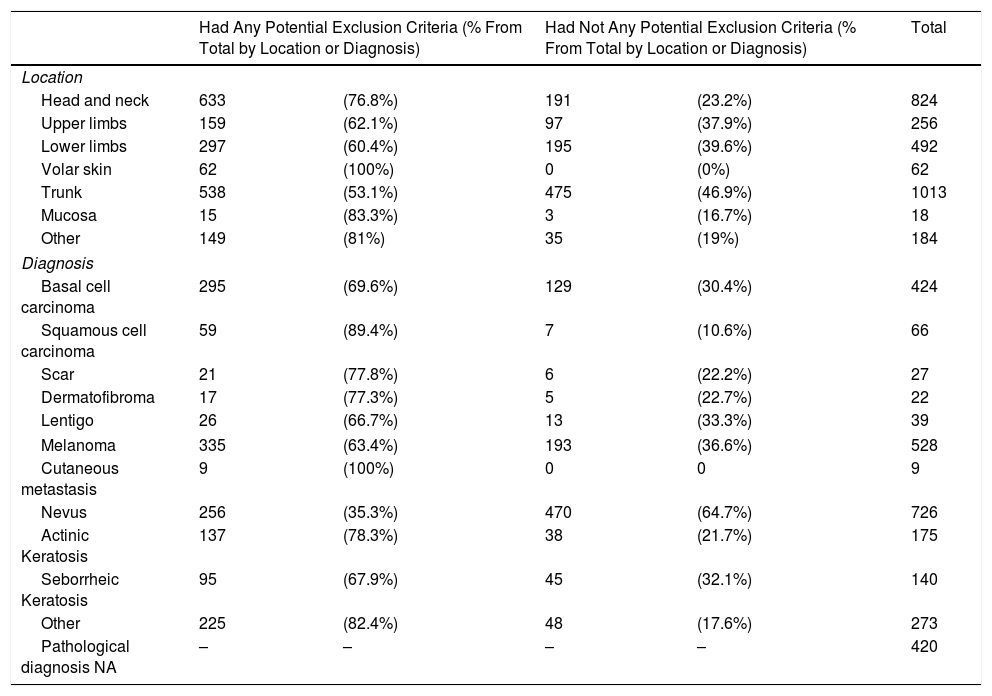
ResultsOut of the 2,849 images from our database, 968 (34%) were selectable as they did not have any potential exclusion criteria for analysis by a ML system. Nevi, melanoma and basal cell carcinoma were the most frequent lesions in our database. Only 64.7% of nevi and 36.6% of melanoma did not have any potential exclusion criteria (Table 1).
Of 528 melanomas, 335 (63.4%) could potentially be excluded. An absence of normal surrounding skin (40.5% of all melanomas) and absence of pigmentation (14.2%) were the most common reasons for exclusion from ML analysis. Other reasons for exclusion are shown in Table 1.
A. Images Chosen for Analysis by ML. Location and Diagnosis.
| Had Any Potential Exclusion Criteria (% From Total by Location or Diagnosis) | Had Not Any Potential Exclusion Criteria (% From Total by Location or Diagnosis) | Total | |||
|---|---|---|---|---|---|
| Location | |||||
| Head and neck | 633 | (76.8%) | 191 | (23.2%) | 824 |
| Upper limbs | 159 | (62.1%) | 97 | (37.9%) | 256 |
| Lower limbs | 297 | (60.4%) | 195 | (39.6%) | 492 |
| Volar skin | 62 | (100%) | 0 | (0%) | 62 |
| Trunk | 538 | (53.1%) | 475 | (46.9%) | 1013 |
| Mucosa | 15 | (83.3%) | 3 | (16.7%) | 18 |
| Other | 149 | (81%) | 35 | (19%) | 184 |
| Diagnosis | |||||
| Basal cell carcinoma | 295 | (69.6%) | 129 | (30.4%) | 424 |
| Squamous cell carcinoma | 59 | (89.4%) | 7 | (10.6%) | 66 |
| Scar | 21 | (77.8%) | 6 | (22.2%) | 27 |
| Dermatofibroma | 17 | (77.3%) | 5 | (22.7%) | 22 |
| Lentigo | 26 | (66.7%) | 13 | (33.3%) | 39 |
| Melanoma | 335 | (63.4%) | 193 | (36.6%) | 528 |
| Cutaneous metastasis | 9 | (100%) | 0 | 0 | 9 |
| Nevus | 256 | (35.3%) | 470 | (64.7%) | 726 |
| Actinic Keratosis | 137 | (78.3%) | 38 | (21.7%) | 175 |
| Seborrheic Keratosis | 95 | (67.9%) | 45 | (32.1%) | 140 |
| Other | 225 | (82.4%) | 48 | (17.6%) | 273 |
| Pathological diagnosis NA | – | – | – | – | 420 |
| B. Reasons for Exclusion | |
|---|---|
| Melanoma | Number of Excluded (% From Total Melanoma) |
| Reasons for exclusion | |
| Absence of pigmentation | 75 (14.2%) |
| Absence of normal surrounding skin | 214 (40.5%) |
| Presence of hair | 28 (5.3%) |
| Metastasis | 29 (5.5%) |
| Location on volar skin | 23 (4.4%) |
| Ulcerated lesion | 19 (3.6%) |
Melanoma accounts for the majority of skin cancer deaths. Early diagnosis and treatment significantly improves its prognosis. The development of an effective screening method is needed and automated image classification by pattern recognition may achieve diagnostic accuracy similar to expert dermatologist.6 However, some limitations have to be overcome. One of these is the exclusion criteria in the selection of skin cancer images. While solely high-quality dermoscopy images were selected from our database, only 34% did not have any potential exclusion criteria for classification by most state-of-the-art ML algorithms. Moreover, 63.4% of our melanomas had at least one of the potential exclusion criteria mentioned above. This considerably decreases diagnostic accuracy and utility of some ML systems. Large lesions are a serious problem for ML algorithms, as they do not fit within the diameter of the majority of dermoscopy lenses, and this renders all the state-of-the-art systems which need to pre-compute lesion segmentation.1 Even if some works have proposed hair detection/removal methods,5 most ML systems’ performance is deteriorated by its presence. Since most dermoscopy datasets for algorithm training don’t include volar skin lesions, the systems trained on these won’t be able to correctly classify acral lesions. Nevertheless, the artificial intelligence community is rapidly moving to overcome these nuances. Yu et al.7 published recently a work where DCNN was used for acral melanoma and nevus classification. In this work we consider the limitations of most but not all ML systems.
Our study shows that the main potential exclusion criteria were the absence of normal surrounding skin and the absence of pigmentation. Many melanomas developed in sun-damaged skin with abnormal surrounding skin, which makes them unsuitable for analysis by most of the current ML systems due to difficulties in lesion border detection.5 Moreover, amelanotic melanoma which accounts for 2%–8% of all melanomas8 cannot yet be diagnosed by most current ML systems. This could be addressed by designing ML systems which are able to work with images which do not contain the entire lesion and increasing the dataset size, selecting a higher number of representative dermoscopy images.
In conclusion, we consider that ML systems, especially those based in the new developments in the deep learning field will not only convert ML into a valuable tool for the dermatologist but also for the general population. However, these systems are able to overcome some limitations to enlarge spectrum of measurable images. It is clear though that researchers are moving forward towards this direction, since some of the exclusion criteria mentioned in this work have already been overcome by recent algorithms included in the ISIC International Symposium.3
Funding/SupportThe study in the Melanoma Unit, Hospital Clínic, Barcelona was supported in part by grants from Fondo de Investigaciones Sanitarias P.I. 12/00840, PI15/00956 and PI15/00716 Spain; by the CIBER de Enfermedades Raras of the Instituto de Salud Carlos III, Spain, co-funded by “Fondo Europeo de Desarrollo Regional (FEDER). Unión Europea. Una manera de hacer Europa”; by the AGAUR 2014_SGR_603 and 2017_SGR_1134 of the Catalan Government, Spain; by a grant from “Fundació La Marató de TV3, 201331-30”, Catalonia, Spain; by the European Commission under the 6th Framework Programme, Contract n°: LSHC-CT-2006-018702 (GenoMEL); by CERCA Programme/Generalitat de Catalunya and by a Research Grant from “Fundación Científica de la Asociación Española Contra el Cáncer” GCB15152978SOEN, Spain. Part of the work was developed at the building Centro Esther Koplowitz, Barcelona.
Conflicts of InterestThe authors declare that they have no conflicts of interest
Thanks to our patients and their families who are the main reason for our studies; to nurses from the Melanoma Unit of Hospital Clínic of Barcelona, Daniel Gabriel, Pablo Iglesias and Maria E Moliner for helping to collect patient data and to Paul Hetherington for helping with English editing and correction of the manuscript.
Please cite this article as: González-Cruz C, Jofre MA, Podlipnik S, Combalia M, Gareau D, Gamboa M, et al. Uso del aprendizaje automático en el diagnóstico del melanoma. Limitaciones por superar. Actas Dermosifiliogr. 2020;111:313–316.